

Next: Phylogenetic Tree
Up: Tools Menu
Previous: Tools Menu
Contents
Subsections
MultiSeq can do both structural and sequence alignments. These options
are available via the Tools menu in MultiSeq.
MultiSeq uses the program STAMP to structurally align protein molecules.
The STAMP algorithm minimizes the  distance between aligned
residues of each molecule by applying globally optimal rigid-body
rotations and translations. Also, note that you can perform alignments
on molecules that are structurally similar. If you try to align proteins
that have no common structures, STAMP will have no means to align them.
If you would like further information about how the alignment occurs,
please refer to the STAMP manual (http://www.compbio.dundee.ac.uk/manuals/stamp.4.2/).
distance between aligned
residues of each molecule by applying globally optimal rigid-body
rotations and translations. Also, note that you can perform alignments
on molecules that are structurally similar. If you try to align proteins
that have no common structures, STAMP will have no means to align them.
If you would like further information about how the alignment occurs,
please refer to the STAMP manual (http://www.compbio.dundee.ac.uk/manuals/stamp.4.2/).
Figure 11:
STAMP Structural Alignment Window
|
|
- Align the following:
- Choose which structures you wish to
align
- Number of passes (npass):
- Whether one or two fits are to
be performed. The idea is that the initial fit can be used with a
conformation biased set of parameters to improve the initial fit
prior to fitting using distance and conformation parameters.
Default NPASS = 2
- Similarity (scanscore):
- Specifies how the Sc value (STAMP
algorithm) is to be calculated. This depends on the particular
application. As a general rule of thumb, use SCANSCORE=6 for large
database scans, when you are scanning with a small domain, and
wishing to find all examples of this domain - even within large
structures. Use SCANSCORE=1 when you wish to obtain a set of
transformations for a set of domains which you know are similar
(and have defined fairly precisely as domains rather than the
larger structure that they may be a part of). Default SCANSCORE =
6
- Comparison residues (scanslide):
- This is the number of
residues that a query sequence is 'slid' along a database sequence
to derive each initial superimposition. Initially, the N-terminus
of the query is aligned to the 1st residue of the databse, once
this fit has been performed and refined, and tested for good
structural similarity, the N-terminus is aligned with the 1+th
position, and the process repeated until the end of the database
sequence has been reached. Default SCANSLIDE = 5
- Slow scan:
- If this box is checked, then the SLOW method of getting
the initial fits for scanning will be used (see the manual for mor information).
Default SLOWSCAN = FALSE
- Defaults:
- resets the STAMP parameters to their original values
Sequence alignment in MultiSeq can be done via ClustalW or MAFFT (if you
have MAFFT locally installed[For installation information see 1.2.3]) (See Fig. 12).
Figure 12:
Sequence Alignment Menu Window
|
|
Once you have decided which program to use, you can
choose from
Multiple Alignment, Profile/Sequence Alignment, or
Profile/Profile Alignment. Once you have chosen the desired type
of alignment, you can set the proper option.
- Multiple Alignment
- Choose which sequences or regions you
wish to align.
- Profile/Sequence Alignment
- This requires certain sequences
to be marked, and they will then be aligned relative to the group that you
specify.
- Profile/Profile Alignment
- To align one entire group with
another entire group, select this option.

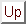


Next: Phylogenetic Tree
Up: Tools Menu
Previous: Tools Menu
Contents
multiseq@scs.uiuc.edu