


Next: Highlight Style
Up: View Menu
Previous: Zoom
Contents
You can choose to color the sequences by a wide range of attributes.
First, you can choose what you want to color by choosing
Apply to All, Group, or Marked. Then, you
can choose the coloring method that you wish to apply. In VMD in Graphics
 Colors in the color scale tab you can choose the overall color scale for
VMD such as RWB and RGB7. Also, Multiseq can
display a color scale for you based on the coloring method selected in VMD(see 9.5).
Colors in the color scale tab you can choose the overall color scale for
VMD such as RWB and RGB7. Also, Multiseq can
display a color scale for you based on the coloring method selected in VMD(see 9.5).
- Add current Selection colors all selected molecules
- Alignment Position colors the sequences based on length where the begining, middle and end
of a sequence will have different colors
- Contact colors the sequence based on the number of contacts or the order of contacts
- Insertions takes the first structure in multiseq and colors sequence insertions based on that sequence
- Mutual Information takes 2 or more align groups and colors them based on mutually shared identities
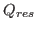 aka Q per residue colors the sequence based on the
contribution from each residue to the overall average Q score. For
more information see 10.3.
aka Q per residue colors the sequence based on the
contribution from each residue to the overall average Q score. For
more information see 10.3.
- Residue Type Colors amino acid residues by whether they are acidic or basic.
- RMSD takes aligned sequences and colors based on the distanced between two
 of aligned residues
of aligned residues
- Sequence Conservation colors residues based on their conservation with all other sequences in Multiseq
- Sequence Entropy colors columns of sequences based on the conservation of sequence
- Sequence Identity method colors each amino acid according to the degree of
conservation within the alignment: blue means highly conserved, wheras
red means very low or no conservation in the default RWB color setting.
- Sequence Similarity colors aligned sequences based on BLOSUM or a custom subsitution matrix
- Signatures colors sequences based on sequence signatures. These options include:
Minimum fraction conserved to be a group signature:, Maximum fraction of signatures allowed in
other groups:, Maximum fraction of gaps allowed in other groups:, Maximum
distance of signature from a conserved block:, Minimum length of conserved block:
- Custom is a way to import custom tcl scripts with specific coloring commands
- Import allows you to import color dat files that had been previously exported using Export Data
- Refresh Color is used if you believe that your newest color change was not implemented



Next: Highlight Style
Up: View Menu
Previous: Zoom
Contents
multiseq@scs.uiuc.edu