For tutorials on NAMD and VMD, which are needed to use MultiSeq click here.
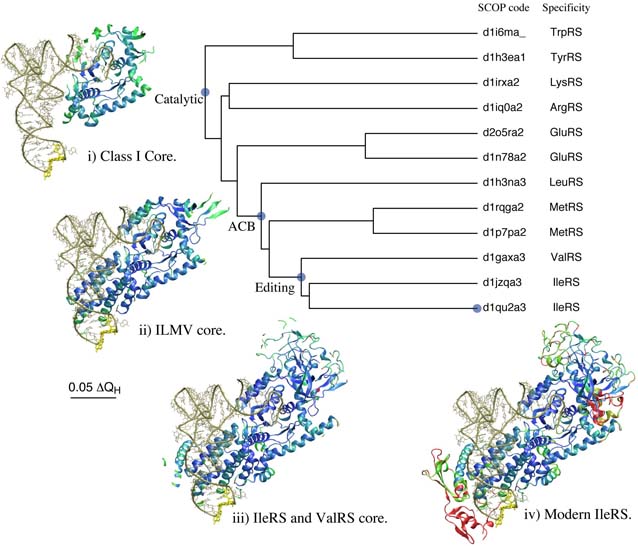
Evolution of Translation: Class-I Aminoacyl-tRNA Synthetases
This tutorial makes use of the MultiSeq bioinformatic analysis environment to explore the evolution of the class I aminoacyl-tRNA synthetases, which 'charge' transfer RNA (tRNA) with the correct amino acid. It is intended to be an introduction to MultiSeq, and no prior knowledge of MultiSeq is required. Topics covered include: BLAST searches, multiple sequence alignments, structural alignments, and distance-based phylogenetic trees. The tutorial is designed such that it can be used by both new and experienced users of VMD, however, it is highly recommended that new users go through the “VMD Molecular Graphics” tutorial in order to gain a working knowledge of the program. This tutorial should take about two hours to complete in its entirety.
The tutorial document is currently available as a PDF (6.7 MB): View Tutorial
(Creation date: 2011-04-15 / Upload date: 2012-06-26)
Files used in the tutorial are available as a compressed file: (361 MB): Download Files
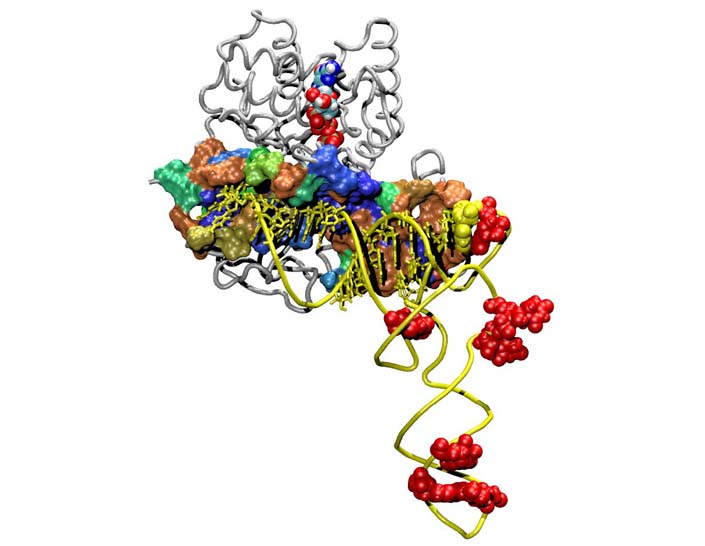
Evolution of Translation: EF-Tu
This tutorial is intended to be a advanced lesson in the MultiSeq bioinformatic analysis environment, and we recommend the user first go through the Class-I Aminoacyl-tRNA synthetases tutorial prior to attempting this one. In it you will explore the evolutionary relationship of the elongation factor Tu, which ferries 'charged' tRNA from the sythetase to the ribosome. Topics covered include: profile-profile alignments, maximum likelihood phylogenetic trees with RAxML, and scripting with MultiSeq. The tutorial is designed such that it can be used by both new and experienced users of VMD, however, it is highly recommended that new users go through the “VMD Molecular Graphics” tutorial in order to gain a working knowledge of the program. This tutorial should take about two hours to complete in its entirety.
The tutorial document is currently available as a PDF (4.9 MB): View Tutorial
(Creation date: 2011-07-05 / Upload date: 2014-09-02)
Files used in the tutorial are available as a compressed file: (285 MB): Download Files
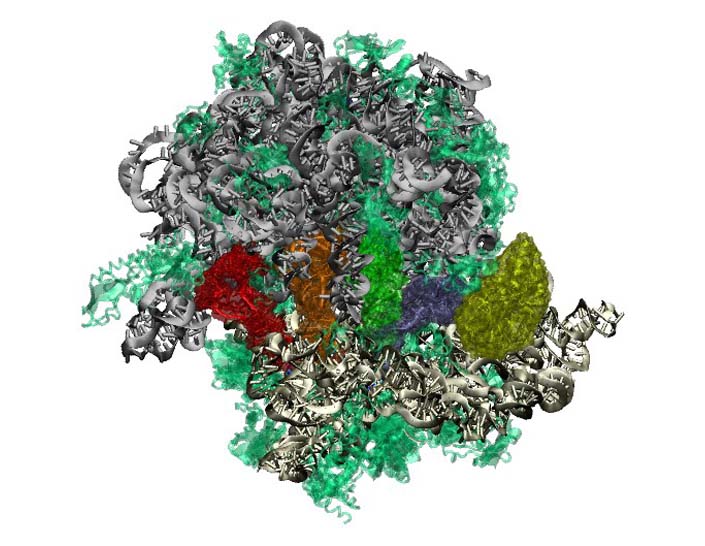
Evolution of Translation: Ribosome
This tutorial leads the reader through major features of the ribosome, the primary translation machinery of the cell. Interactions with the elongation factors, mRNA, and tRNA are explored, as are newly discovered 'sequence signatures' between bacterial and archaeal ribosomes. These signatures constitute much of the evolutionary distance between these two domains of life, and their role in antibiotic resistance is explored. We recommend the reader first complete the Class-I aminoacyl-tRNA synthetase tutorial prior to attempting this one. The tutorial is designed such that it can be used by both new and experienced users of VMD, however, it is highly recommended that new users go through the “VMD Molecular Graphics” tutorial in order to gain a working knowledge of the program. This tutorial should take about three hours to complete in its entirety.
The tutorial document is currently available as a PDF (6 MB): View Tutorial
(Creation date: 2011-04-15 / Upload date: 2015-04-07)
Files used in the tutorial are available as a compressed file: (67 MB): Download Files
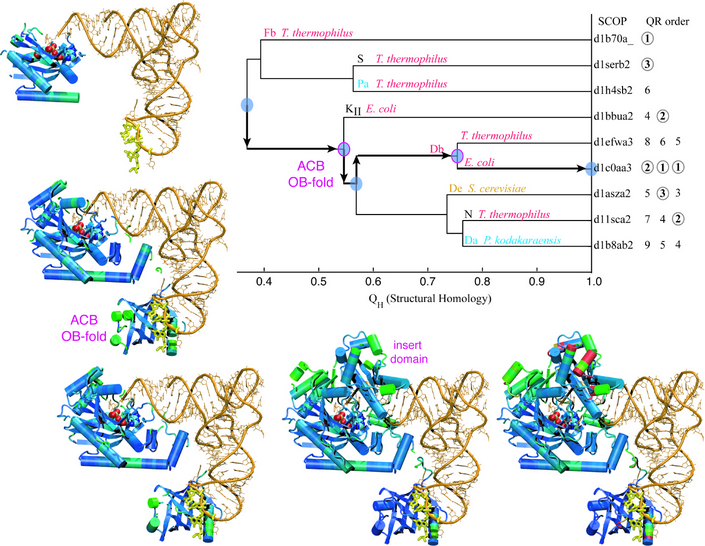
Evolution of Biomolecular Structure: Class-II Aminoacyl-tRNA Synthetases
This tutorial showcases the MultiSeq bioinformatic analysis environment and shows the reader how to combine sequence and structure information into evolutionary profiles. Evolutionary profiles are compact representative sets that can be used, among other things, for gene annotation and homology modeling. The tutorial is designed such that it can be used by both new and experienced users of VMD, however, it is highly recommended that new users go through the “VMD Molecular Graphics” tutorial in order to gain a working knowledge of the program. This tutorial should take about three hours to complete in its entirety.
The tutorial document is currently available as a PDF (2.2 MB): View Tutorial
(Creation date: 2008-02-19 / Upload date: 2012-06-26)
Files used in the tutorial are available as a compressed file: (359 MB): Download Tutorial
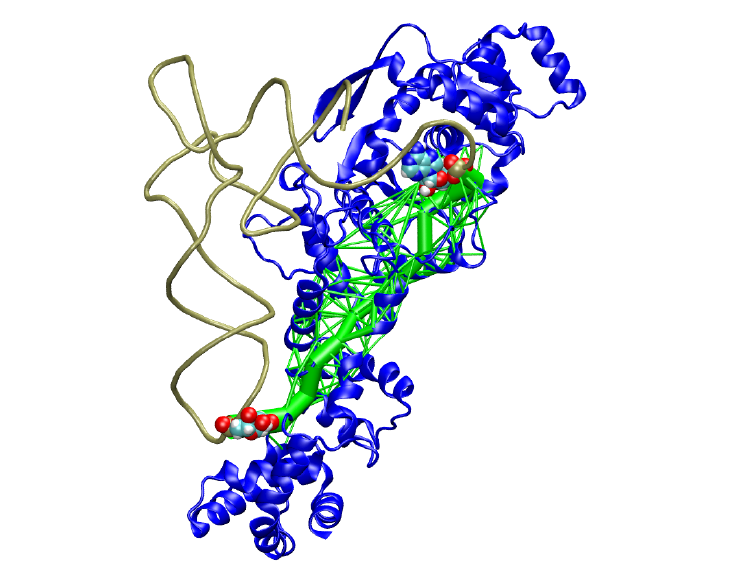
Dynamical Network Analysis
This tutorial introduces several network analysis methods for examining dynamic signaling in biomolecular systems. The NetworkView plugin to VMD is used to display and manipulate representations of the networks projected onto the underlying molecular structures. The tutorial is designed such that it can be used by both new and experienced users of VMD, however, it is highly recommended that new users go through the “VMD Molecular Graphics” tutorial in order to gain a working knowledge of the program. This tutorial should take about two hours to complete in its entirety.
The tutorial document is currently available as a PDF (4 MB): View Tutorial
(Creation date: 2012-06-13 / Upload date: 2012-06-26)
Files used in the tutorial are available as a compressed file: (192 MB): Download Tutorial
Network analysis code is available as a compressed file: (27 MB): Download NetworkTools
NAMD Go tutorial
This tutorial introduces using structure-based modeling in NAMD. Although the tutorial uses a nucleic acid system, it can be easily extended to proteins and other biomolecules as well. The tutorial is designed such that it can be used by both new and experienced users of NAMD, however, it is highly recommended that new users go through the “NAMD” tutorial in order to gain a working knowledge of the program. This tutorial should take between 30 minutes to 5 hours to complete in its entirety.
The tutorial document is currently available as a PDF: View Tutorial
(Creation date: 2013-11-21 / Upload date: 2013-11-21)
Files used in the tutorial are available as a compressed file: Download Tutorial






